We would like to take this opportunity to thank all the present and past members of PharmGKB, our funding agencies, scientific advisors and collaborators, and especially our users, for their continued support and contribution to build this vital resource. PharmGKB serves both basic science investigators as well as clinicians and laboratories. Sustainable long-term support is critically important for us to provide stable, comprehensive, and dependable pharmacogenomic information to our users across the globe.
Thursday, December 15, 2022
PharmGKB selected in the first list of Global Core Biodata Resources
We would like to take this opportunity to thank all the present and past members of PharmGKB, our funding agencies, scientific advisors and collaborators, and especially our users, for their continued support and contribution to build this vital resource. PharmGKB serves both basic science investigators as well as clinicians and laboratories. Sustainable long-term support is critically important for us to provide stable, comprehensive, and dependable pharmacogenomic information to our users across the globe.
Wednesday, November 9, 2022
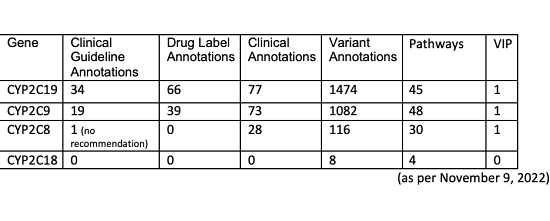
CYP2C18 and knowledge gaps
Thursday, October 20, 2022
PharmCAT Version 2.0 Released
- PharmCAT now includes available DPWG prescribing guidance (as annotated in the PharmGKB DB) in addition to CPIC recommendations. Read more about the genes and drugs included from DPWG and how they are sourced
- PharmCAT report has been redesigned for multiple recommendation sources
- Added functionality for research use only
- CYP2D6 diplotype calling based on SNPs and INDELs from a VCF file (does not include structural variants/CNVs). Warning: Because structural variation and haplotype can’t be determined from VCF, this functionality shouldn’t be used for clinical purposes.
- Partial and combination calls for novel combinations of PGx positions included in the PharmCAT allele definitions. This functionality helps users determine if a sample potentially contains a novel allele.
- Added support for the DPYD genotype based on the lowest activity score as described in CPIC’s fluoropyrimidine guideline (PMID: 29152729) for samples with more than two DPYD variants.
- Extended support of external genotype/phenotype input (see input and output examples and outside call format). This functionality allows users to include genetic test results from other sources in PharmCAT’s report.
- Reworked PharmCAT command line tool/arguments. These changes are not backwards compatible with previous PharmCAT versions.
- VCF Preprocessor updates
- Harmonized the VCF preprocessor command line arguments and flags with PharmCAT
- Unified the output file name patterns of the VCF preprocessor with what PharmCAT uses
- Added a few amenities, e.g., bcftools and bgzip version check, switch to .bgz file suffix for clarity
Tuesday, October 4, 2022
There is no ontology term for Phenoconversion in BioPortal
There are two papers ahead of print in Pharmacogenomics both discussing how important phenoconversion is to consider in the implementation of PGx in clinical practice. Phenoconversion in the PGx context is a drug-drug interaction that impacts a drug metabolizing phenotype such that it mimics the effects of a metabolizer genotype. Yet there was no match for phenoconversion in a search of BioPortal (on 10/3/2022) which has over a thousand biomedical ontologies including MeSH, MedDRA, RxNorm and other ones we use for PGx. PharmGKB does collect drug-drug interaction information from drug labels and publications that can potentially be used in the future to help account for phenoconversion. However, while phenoconversion is a well-known phenomenon, the specifics of how phenoconversion affects patient phenotype, especially on top of genotype, has not been quantified (to our knowledge). This makes it difficult to apply drug-drug interaction information to predict how patient genotype-to-phenotype mapping should be altered by information about concomitant drugs the patient takes when using prescribing guidance from CPIC, DPWG or FDA.
Paper 1: Pharmacogenomics in psychiatry - the challenge of cytochrome P450 enzyme phenoconversion and solutions to assist precision dosing. Mostafa S, Polasek TM, Bousman CA, Müeller DJ, Sheffield LJ, Rembach J, Kirkpatrick CM.Pharmacogenomics. 2022 Sep 28:0. doi: 10.2217/pgs-2022-0104. Online ahead of print. [PMID: 36169629]
This review proposes a model for improved clinical decision support that integrates genomics, co-prescribing information, lifestyle and disease factors into precision dosing. Excerpt from the paper: “In psychiatry, the proposed CDSS (Clinical decision support system) powered by MIPD (model-informed precision dosing) would apply precision dosing of psychotropics by accounting for the influence of genetic variations in CYPs; the presence of CYP phenoconversion; and coexisting lifestyle (smoking), pregnancy or disease (cancer) factors…. In this study, clozapine concentrations were better predicted by MIPD accounting for the CYP1A2 inducing effect in smokers homozygous for the CYP1A2*1F allele. This is an example of where environmental (smoking) and PGx (CYP1A2 genotype) factors were used to optimize the MIPD model, resulting in improved predictions of clozapine plasma concentrations. In principle, this approach can be applied across other psychotropics, especially those with a high risk of toxicity in overdose (e.g., tricyclic antidepressants).”
Paper 2: The importance of phenoconversion when using the CYP2D6 genotype in clinical practice. Cicali EJ, Wiisanen K.Pharmacogenomics. 2022 Sep;23(14):749-752. doi: 10.2217/pgs-2022-0087. Epub 2022 Sep 14. [PMID: 36102178]
This is an editorial with a case study describing a patient with chronic pain taking tramadol (among other medications). The patient is then started on an antidepressant and the pain is no longer relieved even at higher doses. Even though the patient tests as a CYP2D6 normal metabolizer the antidepressant fluoxetine has resulted in phenoconversion and clinically the patient now responds as a CYP2D6 poor metabolizer with respect to tramadol. They discuss options to change the antidepressant or the pain therapies. The authors caution that “CYP2D6 genetic test results should be continually evaluated in the light of concomitant medications throughout a patient’s lifetime.”
Searching PubMed to see the impact of phenoconversion is complicated as this word is also used to describe change or evolution of disease phenotypes, but the results by year tracker shows exponentially increased use. A phenoconversion tag specific for drug interaction related phenoconversion, would help people in PGx research identify the relevant papers.
Maybe phenoconversion could be added as a child term to MedDRA under Drug-drug pharmacokinetic interaction?
Monday, September 26, 2022
New tutorial and walkthrough videos now available
We are pleased to announce the release of our PharmGKB walkthroughs and tutorial videos on YouTube. Users can now view detailed video walkthroughs of each of the main annotation types on PharmGKB as well as a longer video combining resources from across the site. We have also produced a series of tutorial videos to help users learn more about key concepts and issues in pharmacogenomics. These videos range from an introduction to the field to an explanation of the star allele nomenclature system for haplotypes. The videos are freely available on the PharmGKB YouTube channel. Links can also be found on our Educational Resources page.
Thursday, September 22, 2022
PharmGKB and Reactome collaborate on two pathways
PharmGKB has collaborated with Reactome for two new pharmacokinetics pathways, Ribavirin, and Prednisone and Prednisolone available via both formats:
Ribavirin Pathway, PharmacokineticsMonday, August 29, 2022
Study of vitamin K pathways presents potential new warfarin candidates
A review of several ferroptosis related proteins (Vabulas, 2021) discusses some variants of AIFM2. The review mentions a functional study of E156A in the FAD cofactor binding domain that found it impaired anti-ferroptotic activity. This variant is not found in dbSNP. A different amino acid change, E156D (rs1272224219C>A), has not been observed in the ALFA populations that dbSNP reports on, while yet another amino acid change, E156V (rs760393626T>A), is extremely rare (found in 1/121216 alleles). The review lists 2 other potential AIFM2 candidates for functional investigation which are more frequently observed: M135T (mapped by PharmGKB to rs10999147A>G) and D288N (mapped to rs2271694C>T).
(Edited 9/20/22) The Warfarin Pathway, Pharmacodynamics has been updated to include the new candidate gene.
Friday, July 29, 2022
PharmVar GeneFocus paper for SLCO1B1 is published
The PharmVar GeneFocus: SLCO1B1 paper has just been published by Clinical Pharmacology & Therapeutics.
This review provides a general overview of SLCO1B1 as well as a deeper dive into its nomenclature. This GeneFocus covers genetic variability, functional impact, clinical relevance, gene nomenclature before and after PharmVar updates, methods for allele characterization and how the new nomenclature impacts pharmacogenetic testing and interpretation. Specific details of changes to allele definitions can be found on the PharmVar SLCO1B1 page and on the Change Log tab of the SLCO1B1 Allele Definition Table available from PharmGKB.
For more details, please see:
Clin Pharmacol Ther. 2022 Jul 7. doi: 10.1002/cpt.2705.
Laura B. Ramsey, Li Gong, Seung-been Lee, Jonathan B. Wagner, Xujia Zhou, Katrin Sangkuhl, Solomon M. Adams, Robert J. Straka, Philip E. Empey, Erin C. Boone, Teri E. Klein, Mikko Niemi, Andrea Gaedigk.
PMID: 35797228
Friday, July 22, 2022
CYP2A6 now released on PharmVar
PharmVar and PharmGKB are excited to share that CYP2A6 has been transitioned into the PharmVar database. CYP2A6 metabolizes several substates including coumarin, nicotine, aflatoxin B1, nitrosamines, and some pharmaceuticals. Owing to its highly polymorphic nature, CYP2A6 activity varies considerably between individuals. Due to the complex nature of the CYP2A gene locus that contains not only CYP2A6, but also the highly similar CYP2A7 and CYP2A13 genes, CYP2A6 genotype analysis and characterization of allelic variants is not trivial. It is therefore of utmost importance to have up-to-date information regarding sequence variation and star allele (haplotype) definitions to facilitate accurate genetic testing, data interpretation and phenotype prediction in the research and clinical settings.
The PharmVar CYP2A6 gene experts have systematically reviewed and curated all star allele definitions that were previously issued by the CYP450 Nomenclature databases (these were last updated in 2014 and can be accessed through the archive). Some notable changes include:
- Variants and star alleles are now defined using the most current genomic reference sequence
- Several alleles have been merged, revised and/or redesignated (legacy allele designations are cross-referenced)
- Regions used for allele definitions have been updated
- Structural variants including a common conversion at the 3’UTR have been updated to current knowledge and are detailed in the ‘Structural Variation’ document.
Changes made are detailed in the ‘Change Log’ document and other important information about CYP2A6 and the information displayed by PharmVar can be found in the ‘Read Me’ document. All accompanying documents can be accessed at the PharmVar CYP2A6 page at https://www.pharmvar.org/gene/CYP2A6.
Since there are no CPIC guidelines for CYP2A6, the PharmVar CYP2A6 page does not provide information for ‘CPIC clinical function’. The expert panel has, however, compiled a table in the ‘Read Me’ document summarizing function information for selected star alleles.
Lastly, we would like to thank the PharmVar CYP2A6 experts Rachel Tyndale, Alec Langlois, Meghan Chenoweth, Giada Scantamburlo, Charity Nofziger, David Twesigmwe, Rachel Huddart and Andrea Gaedigk for their tireless efforts that made this massive update possible.
Tuesday, June 21, 2022
Clinical Genomics Career Panel webinar series 2022
ClinGen is hosting a Clinical Genomics Career Panel webinar series this summer for individuals interested in career in clinical genomics. Sessions are moderated and panel members will discuss their work and career paths. All are welcome to join!
PharmGKB Acyclovir/Ganciclovir Pathway Published
The PharmGKB Acyclovir/Ganciclovir Pathway has recently been published in the journal Pharmacogenetics and Genomics.
Acyclovir (ACV) and ganciclovir (GCV) are commonly prescribed antivirals to treat infections caused by herpes viruses, varicella-zoster virus or cytomegalovirus (eg. cold sores, shingles and chicken pox, etc.). The pathway, co-developed by Maud Maillard along with other members of the Yang Lab in St. Jude, as well as members of the PharmGKB team, outlines the metabolism, transport, and mechanism of action of ACV and GCV with a view to decipher the existing interpatient variability, and highlights pharmacogenomics implications by the variants of the NUDT15 and ABCC4 genes on ACV and GCV efficacy. Further work is needed to validate these findings and discover other candidates, with the aim of optimizing antiviral therapy.
View the interactive pathway on PharmGKB:
Acyclovir/Ganciclovir Pathway, Pharmacokinetics/Pharmacodynamics
Read our new publication:
PharmGKB summary: acyclovir/ganciclovir pathway
Maud Maillard, Li Gong, Rina Nishii, Jun J Yang, Michelle Whirl-Carrillo, Teri E Klein
Pharmacogenet Genomics. 2022 Jul 1;32(5):201-208. Epub 2022 May 30.
PMID: 35665708
View all pathways on PharmGKB.
Thursday, June 2, 2022
Expansion of pharmacogenetics education agreed as part of lawsuit settlement
Oregon Health & Science University (OHSU) will introduce new educational initiatives on the risks of prescribing the chemotherapy drug capecitabine to patients with DPD deficiency as part of a lawsuit settlement.
The settlement was reached with Joanne McIntyre, whose husband David died as a result of severe capecitabine toxicity. David carried variations in the gene DPYD, which encodes the DPD enzyme. DPD is involved in metabolism of fluoropyrimidine drugs, including capecitabine. Variants in DPYD, such as those that David carried, can inactivate the DPD enzyme, leading to DPD deficiency. Patients with DPD deficiency are unable to properly metabolize capecitabine and other fluoropyrimidines, and are at risk of experiencing severe drug toxicity. In David's case, this toxicity was fatal.
PharmGKB has annotations of several clinical guidelines for capecitabine and DPYD, including those from CPIC and the DPWG. These guidelines uniformly recommend either a dose reduction or selection of an alternative drug in patients with DPD deficiency.
OHSU will hold seminars to educate clinicians on the risks associated with DPD deficiency, how to identify severe capecitabine toxicity in patients and how to administer the antidote. They will also include a module on the topic in their fellowship program and provide a written resource guide to staff in their oncology department. Going forward, patients identified as candidates for capecitabine chemotherapy will be informed of the risks associated with DPD deficiency and, where appropriate, will be offered testing.
We at PharmGKB applaud Joanne's singular dedication to saving patients' lives and OHSU's commitment to implement these changes. Resources on capecitabine pharmacogenomics, including annotations on clinical guidelines for the use of DPYD genotypes in capecitabine prescribing, can be found at the PharmGKB capecitabine drug page.
Thursday, May 19, 2022
Update to PharmGKB Pediatric Summaries - BPCA Drugs
The latest round of PharmGKB’s pediatric drug summaries is now live on PharmGKB pediatric. This release includes summaries for 55 drugs, bringing the total summary count to over 180, now including all drugs on the Best Pharmaceuticals for Children Act (BPCA) priority list in addition to all CPIC guideline drugs.
- Alfentanil
- Amiodarone
- Ampicillin
- Azithromycin
- Bosentan
- Cidofovir
- Ciprofloxacin
- Clindamycin
- Clonidine
- Dexmedetomidine
- Digoxin
- Doxycycline
- Furosemide
- Granisetron
- Griseofulvin
- Heparin
- Hydralazine
- Hydrochlorothiazide
- Hydromorphone
- Hydroxycobalamin
- Hydroxyurea
- Isotretinoin
- Labetalol
- Levofloxacin
- Levothyroxine
- Lidocaine
- Lisinopril
- Lithium
- Lorazepam
- Lurasidone
- Meropenem
- Metformin
- Methylprednisolone
- Midazolam
- Molindone
- Nafcillin
- Nicardipine
- Nifedipine
- Nifurtimox
- Olanzapine
- Pentobarbital
- Piperacillin-Tazobactam
- Pralidoxime
- Prednisolone
- Sertraline
- Sildenafil
- Spironolactone
- Terbutaline
- Timolol
- Topiramate
- Tranexamic Acid
- Valganciclovir
- Vancomycin
- Vecuronium
Monday, May 16, 2022
Response to the American Academy of Pediatrics' Statement "Eliminating Race-Based Medicine"
Looking forward in research, there is an urgent need to direct research efforts towards underserved populations to address the issues of health disparities. Additionally, clinical implementation of pharmacogenomics needs the development of truly race-agnostic dosing guidelines and algorithms.
Michelle Whirl-Carrillo (Co-Principal Investigator and Director of PharmGKB)
Li Gong
Rachel Huddart
Ingrid Keseler
Clarissa J. Klein
Binglan Li
Caroline F. Thorn
Matt W. Wright (Director, Stanford ClinGen)
Mark Woon
Wednesday, May 4, 2022
Ask a Curator for Healthcare Professionals on June 7th
Sign up here for Tuesday June 7th at 12pm EST, 9am PST, 5pm GMT
Want to be notified about future Ask a Curator events? Join our mailing list here.
Thursday, April 21, 2022
What does *1 really mean? And why does that matter?
A Haiku about PGx for National Poetry Month.
Diagnosed star one,
Yet severe toxicity.
What did the lab test?
What does *1 really mean? And why does that matter?
The star alleles of the drug metabolizing enzyme genes are an unusual way of defining variation and are perhaps one of the most misunderstood aspects of pharmacogenomics. Arising from attempts to describe and standardize the molecular basis of different drug phenotypes; debrisoquine poor metabolizer phenotype, etc [PMID: 7773298, PMID: 8807658]. Sometimes authors use the term “wild type”, even for humans, to describe the most common form of the gene or protein in a given population where it displays the expected drug metabolism phenotype. The *1 allele is generally used to denote the absence of the variants tested. However, it is not a stable assignment; a *1/*1 individual only tested at one locus may not have the same genetic sequence as a *1/*1 individual tested for a panel of 10 variants in the same gene. This really matters when evaluating the likelihood of drug phenotype - the "reference" is only as good as the number of variants that were checked. *1 is rather a placeholder, it is the absence of certainty, because even with a panel that covers variants that represent 99% of known variation (in the populations examined so far which are not representative of the full global population) there may still be rare variants with significant impact on protein function that we may not yet have documented. While a *1/*1 may not be at increased risk of toxicity (or other phenotypes), or require a change of dosage immediately, they still have the baseline level of risk and it shouldn’t be a huge surprise if they exhibit an adverse reaction to a drug.
Recent publications of case studies that concluded a lack of involvement of known pharmacogenes without documenting what was tested:
PMID:35180762 - “Acral Skin Rash Caused by Altered Mercaptopurine
Metabolism in Maintenance Therapy for B-Cell
Acute Lymphoblastic Leukemia” excerpt “TPMT and NUDT15 genotype at diagnosis were wildtype alleles and therefore we started with full 6-MP dosing.”
This issue is not limited to pharmacogenes using star allele nomenclature: “5-Fluorouracil Neurotoxicity in the Absence of Dihydropyrimidine Dehydrogenase Deficiency Case Report” [PMID:35419161] excerpt “Our patient tested negative for DPD mutations, but it remains possible she harbored a genetic variant not accounted for in the genetic testing panel. Other known risk factors include mutations in the orotate phosphoribosyltransferase and thymidylate synthase genes… “
We recommend authors always include the list of all variants that were tested, and other features see [PMID:30406943].
Thursday, April 14, 2022
Update to PharmGKB Pediatric Drug Summaries
The third round of PharmGKB's pediatric drug summaries is now live on PharmGKB pediatric. This release includes over 20 additional drugs with a focus on those on the Best Pharmaceuticals for Children Act (BPCA) priority list, including:
- Albendazole
- Amphotericin b
- Artesunate
- Atropine
- Baclofen
- Benznidazole
- Betamethasone
- Canakinumab
- Cefepime
- Ceftazidime
- Chlorproguanil
- Cortisone acetate
- Dopamine
- Eltrombopag
- Epinephrine
- Etomidate
- Fosfomycin
- Glycopyrrolate
- Montelukast
- Nevirapine
- Ritonavir
Wednesday, April 6, 2022
"Ask a Curator" live zoom event
PharmGKB curators will hold a series of live Q&A events over Zoom to help people find and use different aspects of the knowledgebase. We hope this will be a great way to answer questions about PharmGKB and PGx which are specific to users’ individual needs or projects. These events will demonstrate the full extent of the resources available on PharmGKB, as well as details about those resources and how to download, use, and cite data from PharmGKB.
In order to tailor these events for users with similar needs from PharmGKB, the first event will focus on researchers. Future events geared towards educators and clinicians are in the works, as well as events hosted at different times for our global audience. We will also be recording this upcoming event along with future events for those who are unable to attend live. Recordings will be posted on the PharmGKB YouTube channel.
We are asking people to please register in advance with their questions so curators can better focus these events. There will also be the opportunity to ask questions during the event. Events will be limited to 20 participants to allow enough time for everyone’s questions to be answered.
Sign up here for Tuesday April 26th at 12pm EST, 9am PST, 5pm GMT
Can’t make it but want to sign up for a future Ask a Curator? Join our mailing list here.