A recent paper highlights new variants and genes associated with severe fluoropyrimidine-related toxicity in patients who were genotyped as negative for the four DPYD variants the European Medicines Agency recommends testing for.
Online ahead of print in the Human Genomics journal is De Mattia et al "The burden of rare variants in DPYS gene is a novel predictor of the risk of developing severe fluoropyrimidine-related toxicity" [PMID: 37946254].Monday, November 27, 2023
New Fluoropyrimidine Toxicity Variants Reported in DPYS and PPARD
Wednesday, July 12, 2023
Insights on CYP2C19 and phenoconversion
Phenoconversion in the PGx context is a drug-drug interaction that impacts a drug metabolizing phenotype such that it mimics the effects of a metabolizer genotype (see blog from October 2022). Historically much of the discussion on phenoconversion has focused on CYP2D6.
A new paper in Frontiers in Pharmacology investigates the phenoconversion effects of different CYP2C19 inhibitors [PMID:37361233].
Forty donor liver samples were genotyped for CYP2C19 *2, *3 and *17 and the metabolizer phenotypes predicted. Microsomes were assayed with the probe drug s-mephenytoin and then in the presence of strong CYP2C19 inhibitor fluvoxamine, moderate inhibitors omeprazole and voriconazole and weak inhibitor pantoprazole to look at changes in metabolizer status.
Excerpts from paper:
“Our results demonstrate that the outcome of a DDI is dictated by both inhibitor strength and CYP2C19 activity, which is in turn dependent on genotype and non-genetic factors including comorbidities. …
Fluvoxamine, a strong inhibitor of CYP2C19, caused 86% of *1/*17 donors to become phenotypically IM, whereas most of genetically-predicted IMs were converted to a PM phenotype (57%). In accordance with unaltered CYP2C19 activity in patients with gastroesophageal reflux disease taking pantoprazole, weak inhibition by pantoprazole did not induce phenoconversion…
However, the outcomes of DDIs with moderate inhibitors (omeprazole/voriconazole) matched less well to the proposed phenoconversion model by Mostafa et al, which predicted that NMs/IMs convert to a PM phenotype upon moderate inhibition of CYP2C19. In our study, voriconazole, which acts as a moderate CYP2C19 inhibitor, significantly reduced the drug metabolizing capabilities of CYP2C19 by approximately one level (i.e., from a phenotypic NM to a IM). As a result, 40% of the donors (12/30) were converted into IM or PM phenotypes by voriconazole. Though, none of the NMs were converted into PMs, except for one donor who already exhibited impaired CYP2C19 activity in the absence of voriconazole treatment (basal phenoconversion). For omeprazole, phenoconversion into IM or PM phenotypes was even less frequently seen, in only 10% of the donors …
Altogether, our data suggest that CYP2C19 inhibition by moderate inhibitors can result in phenoconversion, but it seems unlikely to result into a PM phenotype for wild-type *1/*1 genotypes.”
There are a number of interesting results and discussion points:
- There is phenoconversion from disease phenotype - namely diabetes.
- The initial concordance for genotype to phenotype with s-mephenytoin was only 40% and the two CYP2C19*17/*17 did not have ultra-rapid UM phenotype (with Vmax in the low normal NM range). The discussion mentions “other (rare) genetic variants within CYP2C19 could also have influenced the mismatch between predicted and observed activities in our study” but it would have been useful to have ruled out *4. The *17/*17 did produce functional mRNA but the *4 is in the start codon and its impact is on translation not transcription [PMID: 9435198].
- There were two *1/*1 outliers with very high UM phenotype that would be interesting to see further genetic analysis of especially given the escitalopram UM CYP2C-haplotype defined by rs2860840T and rs11188059G in [PMID: 33759177].
Overall, this paper shows that while phenoconversion exists for CYP2C19 based on disease status and DDIs, the impact is not a simple downgrading of phenotype (e.g. from IM to PM) that can be applied in a consistent manner across subjects. The authors show that even for strong inhibitors, phenoconversion happens in 40%-86% of subjects with no clear way to predict which subjects would experience phenoconversion and which wouldn’t. More research on how DDIs alter patient-predicted genotype to phenotype is needed to enable better prediction of patient phenotype for PGx drug dosing recommendations.
Monday, May 1, 2023
CYP3A5 genotyping is a more accurate predictor of drug response than race alone
A new paper in Journal of Clinical Pharmacology from a group at Indiana University [PMID:37042314] implemented genotyping for CYP3A5 in a kidney transplant center.
The team used CPIC guidelines for tacrolimus dosing based on CYP3A5 genotype.
Implementation included provider education and clinical decision support in the electronic medical record.
This study reinforces that CYP3A5 genotype is an important predictor of therapeutic tacrolimus trough concentrations. They demonstrate that CYP3A5 normal and intermediate metabolizers had fewer tacrolimus trough concentrations within the desired range post-transplantation and took longer to achieve therapeutic dose than poor metabolizers. While the authors note they were underpowered to measure outcomes, there was a trend towards transplant rejection or all-cause mortality within the first year of transplant based on CYP3A5 metabolizer phenotype.
The paper highlights how, despite the guidelines from CPIC being published in 2015, the FDA label still currently only has language around race-based dose adjustment rather than giving precise guidance based on genotype:
“The FDA drug label recommends higher starting doses in individuals of African ancestry, but only 70% of African Americans are normal/intermediate metabolizers. CYP3A5 normal/intermediate metabolizers are also found among whites and Asians (East Asian and Central/South Asian) at lower frequencies (14% and 44-55%, respectively).”
“Self-reported African American race is more closely associated with CYP3A5 expresser status than other self-reported race categories, but self-reported race is not an accurate surrogate for genotype.”
The discussion is a reminder that pharmacogenomics can play a key role in reducing bias and fulfilling personalized precision medicine.
“Equality and minimization of bias in healthcare has recently become prioritized by healthcare systems as recognition of racial bias has come to the forefront in many non-healthcare aspects of society”
“One dose standard protocols and using race as a surrogate for genotype can both potentiate racial disparities in tacrolimus dosing. Routine CYP3A5 genotyping is a more accurate predictor of drug response than race alone and deemphasizes race as a biological variable in clinical care”
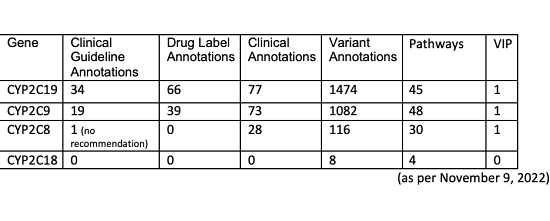
Wednesday, November 9, 2022
CYP2C18 and knowledge gaps
Friday, June 29, 2018
Curators' Favorite Papers
Monday, May 28, 2018
Curators' Favorite Papers
A patient’s diagnosis and medical treatment is frequently guided by where that patient’s laboratory values fall along a range of established “normal” values. A new “Viewpoint” (In the Era of Precision Medicine and Big Data, Who Is Normal?) in the Journal of the American Medical Association (JAMA) raises an important question in the context of laboratory values and precision medicine: if medicine is personalized, to what do we compare an individual’s laboratory values if “normal” is actually relative? The authors consider solutions to ensure that test results “be interpreted in reference to a population of ‘similar’, ‘healthy’ individuals”. For example, the authors propose 1) that longitudinal data on individual outcomes be accessible to researchers to determine whether selected reference values are truly useful 2) that large-scale analyses be carried out across data sets 3) that reference values be tailored to patients and delivered at point of care, and 3) that “computationally derived genetic ancestry” be linked to laboratory test values, so that race is not used as a proxy.
Tuesday, April 24, 2018
Curators' Favorite Papers
Thursday, April 5, 2018
Curator's Favorite Papers
Monday, March 5, 2018
Curators' Favorite Papers
The second paper by Maciel et al. in the journal Neuropsychiatric Disease Treatment (Estimating Cost Savings of Pharmacogenetic Testing for Depression in Real-World Clinical Settings) describes potential cost-savings associated with pharmacogenetic (PGx) testing in patients with depression. The researchers estimated a one-time cost of USD $2,000 for PGx testing and used a published cost-calculator to estimate cost-savings. Data were from a published double-blind, multi-center, randomized clinical trial of 685 adults who had been diagnosed with depression or anxiety. Patients were randomized to PGx-guided treatment or standard of care (control). The study found that those subjects randomized to PGx-guided dosing and prescribing had significant improvements in clinical outcomes as compared to the control group. Using these data, the study calculated that the cost savings for PGx-testing vs standard of care for patients with depression or anxiety totaled USD $3,962 per year after accounting for the one-time cost of PGx testing.
A new meta-analysis of 522 studies that was recently published in The Lancet concluded that at minimum, short-term treatment with anti-depressants was more effective than placebos in treating acute depression. The study made headlines in major media-outlets and found that some anti-depressants worked better than others, which may lead to more interest in PGx-guided dosing.
You can find PGx-guided drug dosing guidelines from the Clinical Pharmacogenetic Implementation Consortium (CPIC) for tricyclic anti-depressants and selective serotonin re-uptake inhibitors on cpicpgx.org. You may also find more information about the following genes and drugs on PharmGKB:
Genes
CYP2D6
CYP2C9
CYP2C19
Drugs
fluoxetine
paroxetine
nortriptyline
desipramine
citalopram
escitalopram
amitriptyline
fluoxetine
Thursday, February 1, 2018
Curators' Favorite Papers
The second paper (Patient understanding of, satisfaction with, and perceived utility of whole-genome sequencing: findings from the MedSeq Project) examines patient-subject understanding of informed consent, satisfaction with disclosure of results, and perception of whole genome sequencing utility (WGS) in cardiology and primary care patients. Patient-subjects were recruited by their cardiologists (already diagnosed with hypertrophic cardiomyopathy or dilated cardiomyopathy) or primary care physicians (PCP) (healthy adults) and were randomized to give family health history (FH) alone or undergo WGS too (FH + WGS). Results were discussed with their respective physicians, who had been counseled in genetics. A majority of subjects (N = 203) were White, non-Hispanic, had completed college and had annual household incomes of over $100K. Most subjects were able to adequately describe the study and demonstrated high genetics and health literacy, as well as an understanding of informed consent and experienced a high level of satisfaction. However, some differences emerged between the FH and the FH + WGS groups: subjects in the FH + WGS group were more likely to report feeling that they had received too much information, and subjects in the FH group reported less satisfaction and more decisional regret than the FH + WGS group. The authors suspect that FH subjects may have been disappointed at not having been randomized to receive WGS results. The primary care group was also more likely to feel that they had received too much information relative to the cardiology group. The authors note that the results are encouraging but suggest that similar efforts in future studies should aim to temper subjects’ expectations regarding WGS and that studies in more diverse cohorts will be necessary.
Thursday, December 28, 2017
Curators' Favorite Papers
Despite the fact that over 100 GPCRs are targeted by approximately 34% of FDA-approved drugs, the frequency of genetic variation of GPCRs is not known according to a study by Hauser et al. from the December issue of Cell (Pharmacogenomics of GPCR Drug Targets). The study evaluates pharmacogenetic (PGx) variation in 108 G-protein coupled receptors (GPCRs) using datasets from the exome aggregation consortium (ExAC) and the 1000 Genomes Project that include over 60,000 individuals and estimates that there is an average of 128 rare and 3.7 common variants per receptor and that 25% of all positions in each GPCR contains a missense variant. In addition approximately 120 of the 60,706 individuals from the dataset harbored loss of function mutations in a GPCR drug target and each GPCR had approximately two duplications and three deletions. The authors support their findings with an analysis of the molecular literature, including data from PharmGKB Clinical Annotations, functional PGx studies of GPCRs on drug response and efficacy and an economic analysis of how incorporation of GPCR PGx could decrease the UKs National Health Service (NHS) financial burden.
Wednesday, November 29, 2017
Curators' Favorite Papers
Monday, October 30, 2017
Curators' Favorite Papers
Dosing and prescribing guidelines involving these and other genes are available on PharmGKB and CPIC.
Thursday, September 28, 2017
Curators' Favorite Papers
Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services by Sissung et al. discusses the experience of the Cleveland Clinic with pharmacogenetic (PGx) implementation as well as how a collaboration between pharmacists and physician-geneticists established an ambulatory PGx clinic to provide testing, interpretation, prescribing recommendations, and patient education. The authors go on to discuss how the implementation of three CPIC guidelines (HLA-B*57:01 and abacavir and HLA-B*15:02 and carbamazepine and TPMT and thiopurines) was managed by pharmacists that incorporated patient data into electronic health records (EHR) who developed clinical decision support tools (CDS) based on CPIC guidelines and included custom pre- and post-test alerts. The selection of which guidelines to implement was based on third-party payment reimbursement for genetic tests, the life-threatening nature of potential adverse events as recognized by the Food and Drug Administration (FDA) in drug labels and physician support.
Pharmacogenomics Implementation at the National Institutes of Health Clinical Center also by Sissung et al. reviews the PGx implementation process at the National Institutes of Health Clinical Center (NIH CC), which uses the recommendations of CPIC guidelines to inform the selection of gene-drug pairs to be implemented. The NIH CC has already implemented PGx testing and prescribing recommendations for HLA-B alleles with allopurinol, abacavir, and carbamazepine, and it is currently in the process of PGx implementation for genes involved in the absorption, distribution, metabolism, or excretion (ADME) of drugs using the DMET genotyping platform. The NIH CC model makes PGx testing, results and recommendations available to all NIH clinicians and also makes test results available to patients and their personal care physicians. The NIH CC expects to cover all CPIC gene-drug pairs within the next five years.
Additional guidelines can be found on the CPIC and PharmGKB and additional information about drugs and genes can also be found on PharmGKB.
Friday, July 7, 2017
Curators' Favorite Papers
Wednesday, April 26, 2017
Curators' Favorite Papers
Authors: Caraballo PJ Bielinski SJ St Sauver JL Weinshilboum RM
The authors of this article from Clinical Pharmacology and Therapeutics, discuss the challenges to successful integration of pharmacogenetic/pharmacogenomic (PGx) clinical decision support (CDS) tools into electronic health and medical records (EMR). They provide examples of medical systems that have already done so and they specifically highlight the success of the Mayo Clinic where CDS tools for eleven PGx genes and nineteen drug-gene interactions have been successfully integrated into EMRs and patient care.
https://www.ncbi.nlm.nih.gov/pubmed/28390138
Celebrating parasites
Authors: Greene CS, Garmire LX, Gilbert JA, Ritchie MD, Hunter LE.
This correspondence to Nature Genetics is a response to a controversial 2016 editorial from the the New England Journal of Medicine in which the term, “research parasites” was coined. The term described those researchers who do secondary analyses of data generated by other researchers. The authors argue that “parasites” serve an important purpose in research and that the critical re-analysis of data is crucial for the practice of science. To honor such research, they announce two inaugural award categories at this year's Pacific Symposium on Biocomputing: the Junior Parasite Award and the Sustained Parasite Award.
https://www.ncbi.nlm.nih.gov/pubmed/28358134